About the Systems Modeling & Data Analytics Core
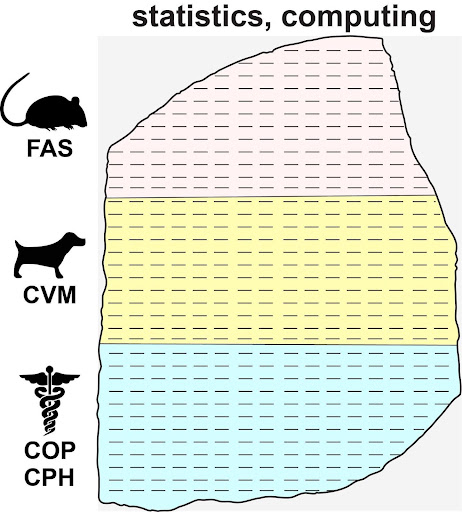
Fig 1. “Rosetta Stone Opportunity”. Common datasets or data-types can serve as a bridge between academic silos at the University of Georgia.
The primary goal of the University of Georgia’s (UGA) Precision One Health (POH) initiative is to provide individual patients with the right medical care at the right time through better treatment and disease-prevention.
The POH rests upon three pillars:
- Pillar 1: Pathogenesis and Diagnostics
- Pillar 2: Therapeutic Intervention
- Pillar 3: Health Promotion and Disease Prevention
Our approach fits the bench-to-bedside–to-community paradigm through an integration of scientific, clinical and community knowledge. For our data core, these knowledgebases are essentially matrices of genomic, environmental and lifestyle data that often overlap across disciplines (Fig. 1). Our methods combine classical approaches in physical modeling, epidemiology and applied statistics with emerging techniques in artificial intelligence and data-science.
The Systems Modeling and Data Analytics Core (SMDA) supports all three POH Pillars as each domain faces similar data-challenges to realize the promise of personalized healthcare (Fig 1). Fortunately, solutions to these challenges have the potential to revolutionize translational research by:
- Increasing the productivity of individual investigators (training)
- Improving communication between disciplines
For example, investigators within the UGA colleges: Arts and Sciences (FAS), Veterinary Medicine (CVM), Pharmacy (COP) and Public Health (CPH), work with datasets on: genomics, therapeutic response, diet and environmental exposure. Better coordination of these data-analytics efforts has the potential to reduce duplicative labor, improve workforce training and enable new research that is clinically relevant and scientifically and statistically rigorous (Fig. 1).
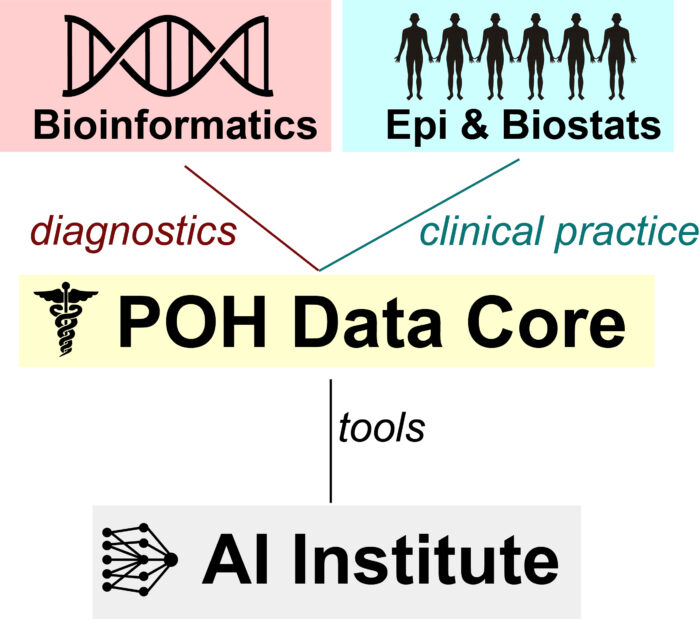
Fig 2. UGA’s Data Ecosystem POH’s data core will serve as an essential link between the Institute of Bioinformatics (IOB), the departments of Epidemiology and Biostatistics and the AI institute (IAI). POH’s leadership and affiliate faculty have appointments within each of these institutes which will prove critical to maintaining lines of communication
The mission of the SMDA Core is to facilitate collaborative research through improved workforce training and communication in the domain of data-analytics and modeling. The SMDA will achieve its mission by building critical infrastructure at two different scales:
- Tactical: Alleviate pain-points for data-driven healthcare research
- Strategic: Build multi-PI working groups that bridge UGA’s discipline- and data-expertise (Fig 2)
The objectives of the SMDA are to facilitate, conduct and disseminate research that advance the primary goals of Precision One Health. This includes but is not limited to:
1.) Facilitate data-analytics training for non-computational investigators:
- pain point: limited workforce availability for data-science/AI
- opportunity: high demand from students for data-analytics training
2.) Facilitate interdisciplinary team research: multi-PI, program project grant, etc.
- pain point: academic silos prevent communication & cause duplicative labor.
- opportunity: recent hires in genomics, data-science, and AI publicly available “big datasets” in every discipline (Fig 1)
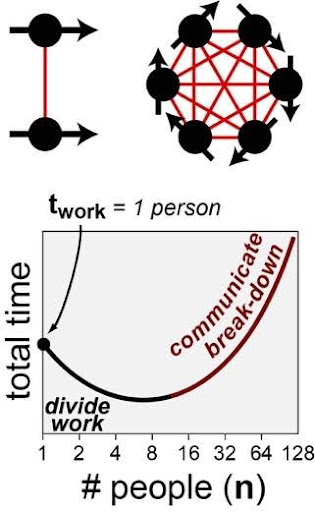
Fig 3. ~8-person teams are often optimal. Paradoxically, very large teams can be less efficient due b>communication complexity (red lines) increasing exponentially with respect to the number of people (sold circles).
Proposed Systems Modeling & Data Analytics Architecture
The proposed architecture for the SMDA core is based on the software-engineering project-management principles described in: “The Mythical Man Month” and modeled after the National Institutes of Health’s (NIH): CTD2 collaborative network of data-driven research centers. A core thesis underlying both initiatives is: information-heavy project operates most efficiently with (Fig 3):
- Small working groups (~8 people/group)
- Efficient communication between working groups (primary goal of leadership and core infrastructure (e.g. website))
The core idea underlying idea these structures is communication (not labor) is the biggest challenge with large team projects (Figure 3 red lines). The NIH’s CTD2 initiative, sought to increase collaboration between large-scale research centers by establishing inter-institution tactical resources:
- Working Groups (WG’s): focused on shared technical challenges. (e.g. genomic, or drug-screening datasets)
- Bridging website (CTD2 Portal): with shared resources for data-challenges (e.g. cleaned datasets and graphic user interfaces)
In parallel with more strategically focused:
- Bi-monthly steering committee meetings
- Yearly conferences
- Bridging website: mapping strategic connections between WG’s
Focused on maintaining communication and establishing strategic priorities. Similar infrastructure is critical for the advancement of data-driven precision health initiatives at UGA. For example, current data-driven infrastructure is being developed within a large data-ecosystem at UGA (Fig 2). A critical role of the SMDA leadership meetings will be to make sure all members and affiliates are kept informed of the (constantly evolving) data-resources that are available to them (Fig 2) and linking individual PI’s with relevant working groups (Fig 3).
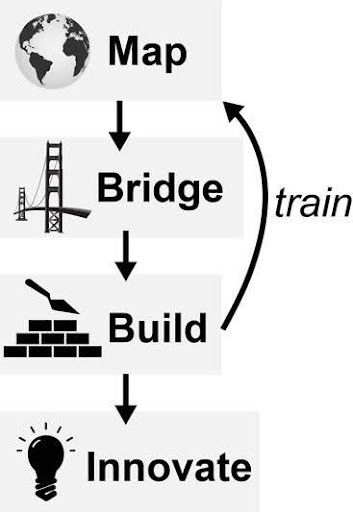
Figure 4. Current Plan to democratize access to data-tools (objective #1) and enable team-based research (objective #2)
Plan to Build Core Foundation for Multi-PI research
Over the next year, we plan to build a collaborative network for data-analytics driven research at UGA using the conceptual plan outlined in Figure 4. First, we will “Map” data-type expertise at UGA to identify tactical overlap across UGA. This first step is critical as currently, data-driven health research is spread across UGA and separated by many traditional silos (Fig 2). Second, we will establish datatype working groups (“Bridge”) to aid coordination of these investigators to reduce duplicative labor and share training resources. These working groups will establish hubs of data-expertise that will be foundational to any large-scale data-driven projects at UGA. For example, genomics research occurs in nearly every biomedical department at UGA and yet, lack of communication causes redundancy and inefficiency in student training and research. The goal of these working groups will be to (1) identify pain-points (2) share resources and (3) coordinate group infrastructure that benefits all members. Third, we will establish Research Question based Working Groups (RQWG’s = “Build”) to link clinical/experimental investigators with data-expertise WG’s. Critical areas of focus will be improving communication and coordination between computational and non-computational investigators. The SMDA website will provide a critical hub for this research by providing tutorial and graphic interfaces for common datasets that are common touchstones across research communities (e.g. The Cancer Genome Atlas (TCGA) for cancer research). This website will provide a centralized resource to enable better coordination between computational and non-computational communities. In addition, these WG’s will establish core training needs for laboratory and clinical researchers who are increasingly becoming responsible for analysis of large datasets. For example, currently PI’s from COP, IAI and CVM are collaborating on a data-analytics course for non-computation investigators that focused on basic data-analytics skills for genomic datasets (PHRM8210). This work has proven foundational to early collaborative research publications and proposals described below.
Interacting and Collaborating with POH Cores:
The SMDA will intentionally look to core and affiliate faculty in other POH cores as collaborators for Research Question Working Groups (Fig 4 “Build”). In addition to POH faculty meetings, point-people will be assigned to companion cores based on joint membership. These point people will provide SMDA with regular updates through monthly email announcements and bi-monthly conference calls.
Interacting and Collaborating with UGA Institutes (Fig 2):
UGA departments and institutes with overlapping interests with our core will be mapped and representatives within core facility will be assigned to each. Each core faculty member will: (1) represent the SMDA interests to each department/institute and (2) keep notes on UGA-initiatives that are relevant to POH. A monthly newsletter will be sent to all core affiliates on new data-initiatives at UGA. This will be initiated during the “Mapping phase” of the work plan described above (Fig 4).
Interacting and Collaborating with External Institutions
As UGA does not (currently) have a medical school, collaborations with academic medical centers will be critical for translational research in humans. Currently we are building a network of with data-analytics PI’s at Emory University and Augusta University to aid collaboration of data-driven research. For example, core director Eugene Douglass, has previously worked Andrey Ivanov and Xiulei Mo (Emory University) through the CTD2 inter-institution network which our core is structurally modeled on. Both investigators have expressed interest in inter-institution collaborations analogous CTD2 which we participated in during our postdoctoral training. This will be initiated during the “Mapping phase” of the work plan described above (Fig 4).
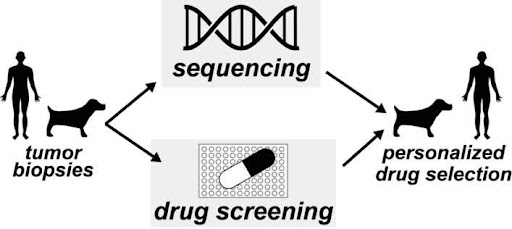
Fig 5. Comparative Medicine Working Group with Focus on integrating clinical knowledge and experimental methods on human and canine-cancers.
Initial Comparative Medicine Working Group
Increasingly, academic medical centers are using drug-screening (in parallel to genomic characterization) to match drugs to patients (Fig. 5). Since 2017, these ex vivo screens have begun to demonstrate superior performance to physicians’ choice and genomically identified targeted therapies. This morning (Feb 15th) the journal Nature published an editorial projecting screening as the future of precision medicine (Nature, 2024 626, 470-473).
Our first research-question working group seeks to build N-of-1 clinical infrastructure at UGA (Fig 5). This work will leverage UGA’s clinical strengths in the College of Veterinary Medicine (CVM) and drug-screening expertise in the College of Pharmacy (COP). This work focuses on canine cancers which provide naturally occurring disease models to better assess clinical efficacy and patient-to-patient variability. Long term, this joint COP-CVM infrastructure could serve as a general model for drug discovery and clinical drug prioritization across other diseases. Critically, this working group is made up of the POH director and two POH core leaders. This leadership team has experience: (1) building a similar infrastructure at Columbia University’s Medical Center, (2) includes two veterinarians with experience in ex vivo tumor culture and canine clinical trials. Our short-term publication/funding plans originate from this RQWG.
12-Month Publication Plan
Currently, our core has three manuscripts in preparation resulting from multi-PI POH-collaborations (Eugene Douglass, Jonathan Mochel, Karin Allenspach, Lorenzo Zapatta):
- A Comprehensive Clinical Dataset and Model for CYP3A4 Drug-Drug Interactions
- Using Physics-based Models to Boost Spatial Transcriptomic Signal
- Preliminary Multi-Tissue Comparison and Characterization of Novel Canine Organoids
12- and 24- Month Funding Plan
During the next year the following funding opportunities have been identified that fit our first RQWG:
- Bladder Cancer Innovation Award: LOI submitted
- NIH R01/R21: Secondary Analysis of Pre-existing Gehomic Data (PAR-23-254)
- NIH R21: ExCITE Award in Cancer Immunology Technology (NOT-CA-24-016)
- NIH R01: Screening Technology Grant (PAR-23-264)
- NIH R01/R21: Biology of Bladder Cancer (PAR-22-218)
- NIH R01: Enhancing Mammalian Models for Translational Research (PAR-23-281)
- NSF EGDE: Enabling Discovery Through Genomics (NSF21546)
After one year we will begin to seek out and apply for program-project level grants through the NIH (P01) or DoD’s Cancer Research Programs (e.g. Breast Cancer Research Programs (BCRP) Breakthrough Level 2 awards).
Overall Timeline and Milestones
Our specific timeline for the plan illustrated in Figure 4 is as follows:
- 2024 Q1: Identify common pain points (e.g. training students, finding collaborators) and map data-expertise across UGA through a combination of survey’s and 1-on-1 meetings with SMDA core and affiliate faculty.
- 2024 Q2: Build preliminary website documenting “UGA Data-Expertise Map” and critical resources for pain-points. Initiate bi-monthly meeting to identify tactical (e.g. website) and strategic priorities (i.e. research). Initiate semi-monthly newsletter to disseminate news to SMDA-faculty from across the UGA-data ecosystem (Fig 2).
- 2024 Q3-4: Establish additional “Research-Question Working Groups (RQWGs)” based on priorities of data- WG’s. Continue development of training infrastructure for core through PHRM8210 course (offered in Fall semesters).
- 2025: With data-expertise foundation set, begin applying to program-project level funding opportunities to be spearheaded by RQWG’s. Leadership will focus on supporting RQWG’s by facilitating communications with data-WG’s and partner institutes within UGA (Fig 2).
Core Faculty
Jonathan Mochel, DVM, PhD, DECVPT, (POH Director)
Department of Pathology, College of Veterinary Medicine
Eugene Douglass, Ph.D., (Core Lead)
College of Pharmacy, Institute of Bioinformatics
Blake Billmyre, Ph.D.,
College of Pharmacy, Department of Infectious Disease
Leigh Anne Clark, Ph.D.
Department of Pathology, College of Veterinary Medicine
Lok Raj Joshi, Ph.D.
Poultry Diagnostic and Research Center
Tatum Mortimer, Ph.D.
Dept of Population Health, College of Veterinary Medicine
Tao Tao Wu, Ph.D.
College of Engineering, Institute of Bioinformatics
Niying Li, Ph.D.
Clinical and Administrative Pharmacy, College of Pharmacy
Lorenzo Zapata, Ph.D.
Clinical and Administrative Pharmacy, College of Pharmacy
Affiliate Faculty
Artificial Intelligence Faculty
Chao Huang
Chao’s research interests mainly focus on statistical learning of large-scale biomedical data including clinical, imaging, and genomic data. His research aims to develop novel statistical methods and machine learning (deep learning) algorithms for analyzing data with complex structures, including high dimensional data, functional data, manifold data, and data with heterogeneity. These statistical methods and computational tools can help us understand the disease progression and improve clinical trials for treatment and early prevention. Some projects that he is currently working on are big data integration, manifold data analysis, functional data analysis, imaging heterogeneity, imaging genetics, and deep learning.
Tianming Liu
Dr. Tianming Liu is a Distinguished Research Professor and a Full Professor of Computer Science at The University of Georgia. Dr. Liu’s research interests are brain imaging, computational neuroscience, brain-inspired artificial intelligence, artificial general intelligence, and data science/AI applications. Dr. Liu has published 400+ research papers on these topics, his Google citation is over 16,000+, and his H-index is 66. Dr. Liu is the recipient of NIH Career Award and NSF CAREER Award. Dr. Liu serves on the editorial boards of multiple international journals including IEEE Transactions on Neural Networks and Learning Systems, IEEE Transactions on Medical Imaging, Medical Image Analysis, IEEE Transactions on Cognitive and Developmental Systems, IEEE/ACM Transactions on Computational Biology and Bioinformatics, IEEE Reviews in Biomedical Engineering, and IEEE Journal of Biomedical and Health Informatics. Dr. Liu is a Fellow of AIMBE (American Institute of Medical and Biological Engineering) and was the General Chair of MICCAI (Medical Image Computing and Computer Assisted Intervention) 2019.
Khaled Rasheed
Dr. Khaled Rasheed is a Professor at the School of Computing, University of Georgia. He is also the Interim Executive Director of the UGA Institute for Artificial Intelligence. He received his Ph.D. from Rutgers University in January 1998. Dr. Rasheed is also the founder and director of the Evolutionary Computation & Machine Learning (ECML) Lab. Research in the ECML Lab is centered around Genetic and Evolutionary Algorithms, Machine Learning and the intersection/ cross-fertilization of the two fields. We conduct research in genetic algorithm methodologies and applications in science and engineering. We also develop, apply and analyze machine learning approaches for numerous Bioinformatics and computational biology domains. We have ongoing research in Human Activity Recognition using Deep Learning and Transfer Learning, using time-series techniques for Weather and Biomass Yield Prediction for Sustainability. We have recently conducted research in Identification of Cancer-Causing Mutations in the Human Genome and using Machine Learning Methods for Alzheimer Disease Detection and Prediction.
His research interests include artificial intelligence, genetic algorithms, design optimization, Bioinformatics, and machine learning. He has served on the program committees of several conferences including the Genetic and Evolutionary Computation Conference, the Congress on Evolutionary Computation, Parallel Problem Solving from Nature and the International Conference on Machine Learning.
Jin Lu
Dr. Jin Lu is an Assistant Professor at the School of Computing, University of Georgia. His research spans machine learning, data mining, optimization, smart mobility, biomedical informatics, and health informatics. Dr. Lu is especially keen on developing high-performance, interpretable algorithms to address critical issues in science and engineering. With over a decade of experience in Data Science, Machine Learning, Optimization, and Intelligent Systems, his contributions have been recognized in leading journals and conferences such as ICML, NeurIPS, IEEE MASS, BMC Journal of Systems Biology, IEEE Transactions on Big Data, Ubicomp, IEEE UIC, and many more.
Genomic Diagnostics Faculty
Kaixiong Ye
Kaixiong (Calvin) received his Bachelor's degree in Biology from Wuhan University. He worked briefly at the Beijing Genomics Institute (BGI) as a bioinformatician before he joined graduate school at Cornell University, where he earned his Ph.D. degree in Nutritional Genomics. He received his postdoctoral training in Computational Biology also at Cornell. Kaixiong started his own research group at UGA in 2018. Kaixiong’s research has been driven by his curiosity of the evolution and diversity of life, and by his passion for improving human health. Research in the Ye lab strives to understand gene- environment interactions that make each one of us unique in our nutritional requirements and risks of metabolic diseases. It employs computational and experimental methodologies from Evolutionary and
Quantitative Genetics, Molecular Genetics, and Nutritional Sciences. The nutrients of special interest in the Ye lab are the omega-3 and omega-6 fatty acids (e.g., fish oil supplements).
Srimanti Duttagupta
Dr. Srimanti Duttagupta is an Assistant Research Scientist in Environmental Geoscience at the Department of Geology at the University of Georgia. Her interdisciplinary research interests span environmental and public health and epidemiology. Her work integrates environmental science and public health, aiming to address pressing issues related to environmental contaminants and their impact on ecosystems and human health. She holds a Ph.D. in Environmental Science and Engineering
from the Indian Institute of Technology Kharagpur, where she conducted pioneering research on the occurrence, fate, and transport of persistent organic pollutants in the Western Bengal Basin. Her research also includes untargeted approaches to evaluate anthropogenic and geogenic organic contaminants and their environmental transport in the ecosystem. By leveraging extensive data analysis, she enhances the detection and understanding of contaminants, facilitating more accurate and comprehensive environmental monitoring and risk assessment. She voluntarily serves as an adjunct assistant professor at the School of Public Health at San Diego State University.
Shaying Zhao
I have focused on dog-human comparative genomics and oncology research since 2005. I am the first person to propose and use a novel dog-human comparison strategy for cancer driver-passenger discrimination, a central aim of cancer research. Funded by the National Cancer Institute, the American Cancer Society and the Georgia Cancer Coalition, work from my lab has shown a strong dog-human molecular homology for histologically matched cancer types/subtypes for mammary cancer, colorectal cancer, and head and neck squamous cell carcinoma. My lab has successfully used this novel dog-human comparison strategy for cancer driver-passenger discrimination for amplified or deleted genes in human colorectal cancer (Oncogene 33:814-22). My lab has published the first comprehensive characterization of canine mammary cancers with whole genome sequencing (WGS), whole exome sequencing (WES), and RNA-seq analysis (Cancer Res. 74:5045-56). The same is true for canine head & neck cancer (PLOS Genetics, 11(6):e1005277), colorectal cancer and extreme intestinal polyposis that resembles human familial adenomatous polyposis (FAP). Funded by the American Kennel Club Canine Health Foundation and the English Cocker Spaniel Club of Northern California, my lab is searching for germline mutations that predispose dogs to cancer development, and to improve canine gene annotation by RNA-seq.
Natarajan Kannan
Dr. Kannan is a Professor of Biochemistry and Molecular Biology and the Institute of Bioinformatics at the University of Georgia, USA. His interdisciplinary research group leverages advances in artificial intelligence, bioinformatics, and systems biochemistry to map the complex relationships connecting sequence and function in biomedically important protein families such as protein kinases, glycosyltransferases and ion-channels. Dr. Kannan has published nearly 100 peer-reviewed articles with over 6000 citations and an h-index 41. He has trained over 30 undergraduate, graduate, and post-graduate trainees, junior scientists, and young investigators and has received multiple state and national awards, including the Georgia Coalition Distinguished Scholar Award, National Science Foundation CAREER award, NIH Maximizing Research Investigator Award, and UGA Creative Research Award. See http://esbg.bmb.uga.edu/ for more details.
Aditya Mishra
Dr. Aditya Mishra is an environmental AI cluster hire faculty in the Department of Statistics. Prior to joining the department, Dr. Mishra was a computational/data scientist at the Platform for Innovative Microbiome and Translational Research at the University of Texas MD Anderson Cancer Center. At MD Anderson, Aditya was involved in developing and applying statistical methods to understand the role of the human microbiome in cancer onset, progression, and response to therapy. Aditya’s research mainly focuses on developing statistical methods and computational tools using the framework of High-dimensional statistics, Multivariate analysis, Reduced-rank regression, Regularization, Robust statistics, Multi-omics, Computational biology, Causal inference and Variational inference. He has extensive interdisciplinary research experience in a variety of fields, including microbiome data analysis, genomics, cancer genomics, public health, ocean microbiology, and dietary intervention study. As a part of environmental AI cluster hire faculty, Aditya will continue to understand and investigate the role of microbiome various context including cancer genomics, marine science, soil science and agriculture science.
Kosuke Funato
Dr. Funato received a PhD in biochemistry and biophysics at the University of Tokyo and completed the Undergraduate Program for Bioinformatics and Systems Biology (equivalent to a minor). He conducted postdoctoral work at Memorial Sloan Kettering Cancer Center under the supervision of Dr. Viviane Tabar. In 2021, he started his own lab at the Center for Molecular Medicine at the University of Georgia. His research focuses on malignant brain tumor, particularly pediatric glioblastoma. His team has been utilizing human pluripotent stem cells (including ES cells and iPS cells) as a modeling platform to dissect the molecular mechanisms underlying the formation and progression of malignant brain tumors.
Physics Based Modeling Faculty
Mellissa Hallow
Bio coming soon.
Robin Southwood
I am a clinical associate professor at the University of Georgia College of Pharmacy. I lead instruction on diabetes related topics in our curriculum. I also work with the inpatient diabetes education team at St. Mary’s of Athens hospital.
Virginia Fleming
Bio coming soon.
Andreas Handel
I am Professor and Associate Department Head in the Department of Epidemiology and Biostatistics in the College of Public Health at the University of Georgia. I have adjunct appointments in UGA’s School of Ecology and Department of Infectious Diseases in the College of Veterinary Medicine, as well as the Department of Epidemiology at the Rollins School of Public Health at Emory University. I am also a Member of UGA’s Institute of Bioinformatics and Faculty of Infectious Diseases.
I work on data analytics and modeling of infectious diseases, mainly influenza, tuberculosis and norovirus. Lately, I have also worked a lot on COVID-19. I uses mathematical models, computational simulations and statistical analysis to understand the dynamics of pathogens on different spatial and temporal scales. I work both on the within-host level (the immunology, virology, microbiology scale) and the population level (the epidemiology, ecology, evolution scale). The ultimate goal of my work is to help design better intervention and control strategies against infectious diseases, both for individual patients and on the population level.
Clinical Practice Faculty
Richard Lamb
Bio coming soon.
Matthew Schmidt
Matthew Schmidt, Ph.D., is Associate Professor at the University of Georgia (UGA) in the Learning, Design, and Technology department. His primary research interest includes design and development of innovative educational courseware and computer software with a particular focus on individuals with disabilities, their families, and their providers. His secondary research interests include learning in extended reality (inclusive of virtual reality, augmented reality, and mixed reality) and Learning Experience Design.
Andrea Sikora
Andrea Sikora, PharmD, MSCR, BCCCP, FCCM, FCCP is a Clinical Associate Professor at the University of Georgia (UGA) College of Pharmacy. She practices as a critical care pharmacist at AU Medical Center in Augusta, Georgia. She graduated from UGA College of Pharmacy, completed her PGY1 Pharmacy Residency and PGY2 Critical Care Pharmacy Residency at the University of North Carolina (UNC) Medical Center in Chapel Hill, NC, and completed her Master of Science in Clinical Research from Emory University. She has received several federal grants evaluating applications of artificial intelligence for optimized medication use in the intensive care unit (ICU) and is an author of the book Pay It Forward: A Map to Mentorship in Medicine.
Susan Smith
Susan E. Smith, PharmD, BCCCP, FCCM is a Clinical Associate Professor at the University of Georgia College of Pharmacy in the Department of Clinical and Administrative Pharmacy and an Adjunct Clinical Assistant Professor at the AU/UGA Medical Partnership. She maintains an active practice site as a Critical Care Pharmacist at Piedmont Athens Regional where she provides clinical rotations and research training to pharmacy students, medical students, and pharmacy residents. Dr. Smith is a founding member and chair of the University of Georgia Critical Care Collaborative (UGAC3), which is committed to fostering critical care research and education across the state of Georgia. She has a robust research program focused on optimizing critical care pharmacy practice models and the pharmaceutical care of critically ill patients.
Daniel Chastain
Daniel Chastain is a Clinical Associate Professor at the University of Georgia College of Pharmacy on the Southwest Georgia Clinical Campus and serves as the Infectious Diseases Pharmacist for Phoebe Putney Memorial Hospital in Albany, Georgia. He received his PharmD from South University in Savannah, in 2012, and subsequently completed a PGY-1 at Tallahassee Memorial Healthcare and a PGY-2 in infectious diseases at the University of Mississippi Medical Center. He is recognized as an HIV Pharmacist by the American Academy of HIV Medicine and is a board-certified infectious diseases pharmacist. His teaching, research, and patient care activities focus on infectious diseases pharmacotherapy with emphasis on invasive fungal infections, HIV, and vulnerable populations.
Henry Young
Henry N. Young, PhD, FAPhA received his Doctor of Philosophy degree in Pharmaceutical Sciences – Pharmacy Health Care Administration from the University of Florida and completed a postdoctoral fellowship at the University of California, Davis in the Center for Health Services Research in Primary Care. Dr. Young joined the University of Georgia College of Pharmacy faculty in July 2013. He is the Department Head and Kroger Professor in the Department of Clinical and Administrative Pharmacy. Dr. Young is a behavioral and social scientist with expertise in health services research. His research interests include studying medication use, health communication, and health outcomes, focusing specifically on minority and underserved populations.
UGA Infrastructure Faculty
Guy Cormier
Dr. Guy Cormier is UGA’s Director of Research Computing, directing the Georgia Advanced Computing Resource Center since late 2012. Dr. Cormier brings over 25 years of experience in the implementation and management of High Performance Computing, AI, data-intensive and scientific visualization environments.
Dr. Cormier graduated in 1993 with a Ph.D. in Chemistry from Concordia University, Canada, with a focus in computational chemistry and laser spectroscopy. This was followed by a one-year post-doctoral position at the Chemistry Department of the Università di Padova, Italy, followed by three years as Research Associate at the Chemistry Department of the University of Cambridge, U.K. In late 1997 he joined the University of Puerto Rico System as the Founding Director of UPR’s High Performance Computing Facility, bringing HPC and scientific visualization resources to researchers of the three graduate research campuses of the UPR. From 2006 to 2009, he was the Chief Information Officer of the Virginia Bioinformatics Institute at Virginia Tech. He then joined the Department of Biological Sciences at VT as a Visiting Researcher providing HPC expertise in a number of large-scale collaborative research projects, leaving in 2012 to join UGA.
Walter Lorenz
Bio coming soon.
Fred Maier
Dr. Maier is the Associate Director of UGA’s Institute for Artificial Intelligence, with a research focus in logic and artificial intelligence, particularly knowledge representation and nonclassical reasoning. Before earning a MS in artificial intelligence and a PhD in computer science, he studied philosophy at Tulane University in New Orleans.